
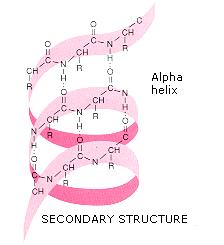
The α-helix is tightly packed there is almost no free space within the helix. The general formula for the rotation angle Ω per residue of any polypeptide helix with trans isomers is given by the equation For comparison, the sum of the dihedral angles for aģ 10 helix is roughly -75°, whereas that for the π-helix is roughly -130°. Consequently, α-helical dihedral angles generally fall on a diagonal stripe on the Ramachandran plot (of slope -1), ranging from (-90°, -15°) to (-35°, -70°). More generally, they adopt dihedral angles such that the ψ dihedral angle of one residue and the φ dihedral angle of the next residue sum to roughly -105°. Residues in α-helices typically adopt backbone (φ, ψ) dihedral angles around (-60°, -45°). Transient helices (sometimes called δ-helices) have also been reported as intermediates in molecular dynamics simulations of α-helical folding. These alternative helices are relatively rare, although the 3 10 helix is often found at the ends of α-helices, "closing" them off. Similar structures include the 3 10 helix ( hydrogen bonding) and the π-helix ( hydrogen bonding). Most importantly, the N-H group of an amino acid forms a hydrogen bond with the C=O group of the amino acid four residues earlier this repeated hydrogen bonding defines an α-helix. Each amino acid corresponds to a 100° turn in the helix (i.e., the helix has 3.6 residues per turn), and a translation of 1.5 Å (= 0.15 nm) along the helical axis. The amino acids in an α helix are arranged in a right-handed helical structure, 5.4 Å (= 0.54 nm) wide. Pauling then worked with Corey and Branson to confirm his model before publication. After a few attempts, he produced a model with physically plausible hydrogen bonds. Being bored, he drew a polypeptide chain of roughly correct dimensions on a strip of paper and folded it into a helix, being careful to maintain the planar peptide bonds. The pivotal moment came in January 1948, when Pauling caught a cold and went to bed. Two key developments in the modeling of the modern α-helix were (1) the correct bond geometry, thanks to the crystal structure determinations of amino acids and peptides and Pauling's prediction of planar peptide bonds and (2) the relinquishing of the assumption of an integral number of residues per turn of the helix. Taylor, Maurice Huggins and Bragg and collaborators to propose models of keratin that resemble the modern α-helix. Interestingly, Neurath's paper and Astbury's data inspired H. Hans Neurath was the first to show that Astbury's models could not be correct in detail, because they involved clashes of atoms. Although incorrect in their details, Astbury's models of these forms were correct in essence and correspond to modern elements of secondary structure, the α-helix and the β-strand (Astbury's nomenclature was kept), which were developed by Linus Pauling, Robert Corey and Herman Branson in 1951 (see below). Astbury proposed that (1) the unstretched protein molecules formed a helix (which he called the α-form) and (2) the stretching caused the helix to uncoil, forming an extended state (which he called the β-form).

The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ~5.1 Å (= 0.51 nm). In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching.


 0 kommentar(er)
0 kommentar(er)
